Research Centre for Computational Health
About us
The Research Centre for Computational Health addresses problems in medicine and biology using data-driven and mechanistic modeling. Important tools are machine learning for image and signal analysis, graphical networks, parameter estimation for differential equation systems and physiological simulation.
Biomedical Simulation
The research group specializes in modeling biological and medical systems. New approaches are developed to simulate physiological processes and to predict pathological changes. In particular, in-depth knowledge of biological/physiological processes is incorporated into multi-physics simulations.
The group develops algorithms for parameter and uncertainty estimation of physically motivated stochastic models. In particular, machine learning methods are combined with Bayesian modeling for dimensionality reduction. These methods are widely used in medicine and life sciences.
Group leader: Prof. Dr. Sven Hirsch | Learn more about the research group Biomedical Simulation
Medical Image Analysis
The research group applies machine learning techniques to interpret medical image data. This way, features are extracted for the characterization of disease patterns and for use as diagnostic markers. Of particular interest are the radiomic and morphological analysis of diagnostic medical imaging data. The group pursues the goal of establishing reproducible, image-based biomarkers by means of explainable artificial intelligence and ensuring their clinical utility.
Medical Data Modelling
The research group uses statistical and machine learning methods to model and uncover causal relations in medical data, especially to study pathophysiological processes. Categorial patient data and imaging date, e.g. magnetic resonance imaging, are processed to extract clinical knowledge.
Biosignal Analysis & Digital Health
The research group studies data from wearables and biosensors using time series analysis and combines them with biological-physical models to robustly characterize physiological systems. These data sources are used for Patient Reported Outcomes in clinical practice and for the further development of patient-centered medicine.
Group leader: Dr. Samuel Wehrli | Learn more about the research group Biosignal Analysis & Digital Health
Software & Code
We provide code and projects developed by the Research Centre for Computational Health on our GitHub organization page. Our repositories cover various topics, including algorithm development, data analysis, and computational modeling. By openly sharing our work, we aim to encourage collaboration and contribute to the wider scientific community. Embracing open science ensures that our research is transparent, reproducible, and accessible, which promotes wider dissemination and accelerates scientific progress. We welcome you to explore our projects, access our source code, and connect with us on GitHub.
Teaching Activities
The courses offered by the centre of Computational Health cover topics in mathematical and physical modeling. At Bachelor level it is responsible for the specialization Digital Health. The specialization teaches physics and mathematics for other BSc programs at the department. In the MSc Applied Computational Life Sciences and the PhD program Data Science advanced courses in modeling of complex systems are taught.
Projects
Unfortunately, no list of projects can be displayed here at the moment. Until the list is available again, the project search on the ZHAW homepage can be used.
Publications
-
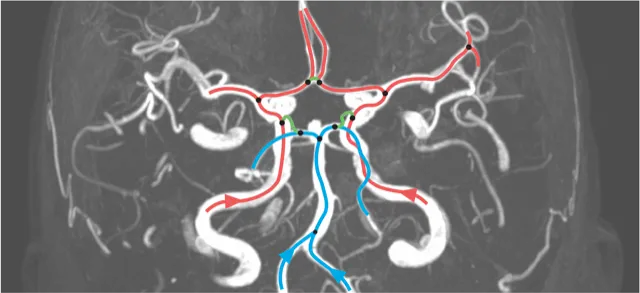
Juchler, Norman; Schilling, Sabine; Bijlenga, Philippe; Morel, Sandrine; Rüfenacht, Daniel; Kurtcuoglu, Vartan; Hirsch, Sven,
2020.
Shape irregularity of the intracranial aneurysm lumen exhibits diagnostic value.
Acta Neurochirurgica.
162(9), pp. 2261-2270.
Available from: https://doi.org/10.1007/s00701-020-04428-0
-
Albert, Carlo; Gaia, Filippo; Ulzega, Simone,
2020.
Can stochastic resonance explain the amplification of planetary tidal forcing?.
In:
EGU General Assembly 2020, Online, 4-8 May 2020.
Available from: https://presentations.copernicus.org/EGU2020/EGU2020-15185_presentation.pdf
-
Juchler, Norman; Schilling, Sabine; Glüge, Stefan; Bijlenga, Philippe; Rüfenacht, Daniel; Kurtcuoglu, Vartan; Hirsch, Sven,
2020.
Computer Methods in Biomechanics and Biomedical Engineering : Imaging & Visualization.
8(5), pp. 538-546.
Available from: https://doi.org/10.1080/21681163.2020.1728579
-
Detmer, Felicitas J.; Mut, Fernando; Slawski, Martin; Hirsch, Sven; Bijlenga, Philippe; Cebral, Juan R.,
2020.
Acta Neurochirurgica.
162(3), pp. 553-566.
Available from: https://doi.org/10.1007/s00701-020-04234-8
-
Watanabe, Kazuhiro; Anzai, Hitomi; Juchler, Norman; Hirsch, Sven; Ohta, Makoto,
2020.
In:
International Mechanical Engineering Congress and Exposition : Volume 3 - biomedical and biotechnology engineering.
2019 International Mechanical Engineering Congress and Exposition, IMECE2019, Salt Lake City, Utah, USA, 11-14 November 2019.
The American Society of Mechanical Engineers.
Available from: https://doi.org/10.1115/IMECE2019-11125