Research Projects
Technologies
Algorithm-assisted enzyme engineering
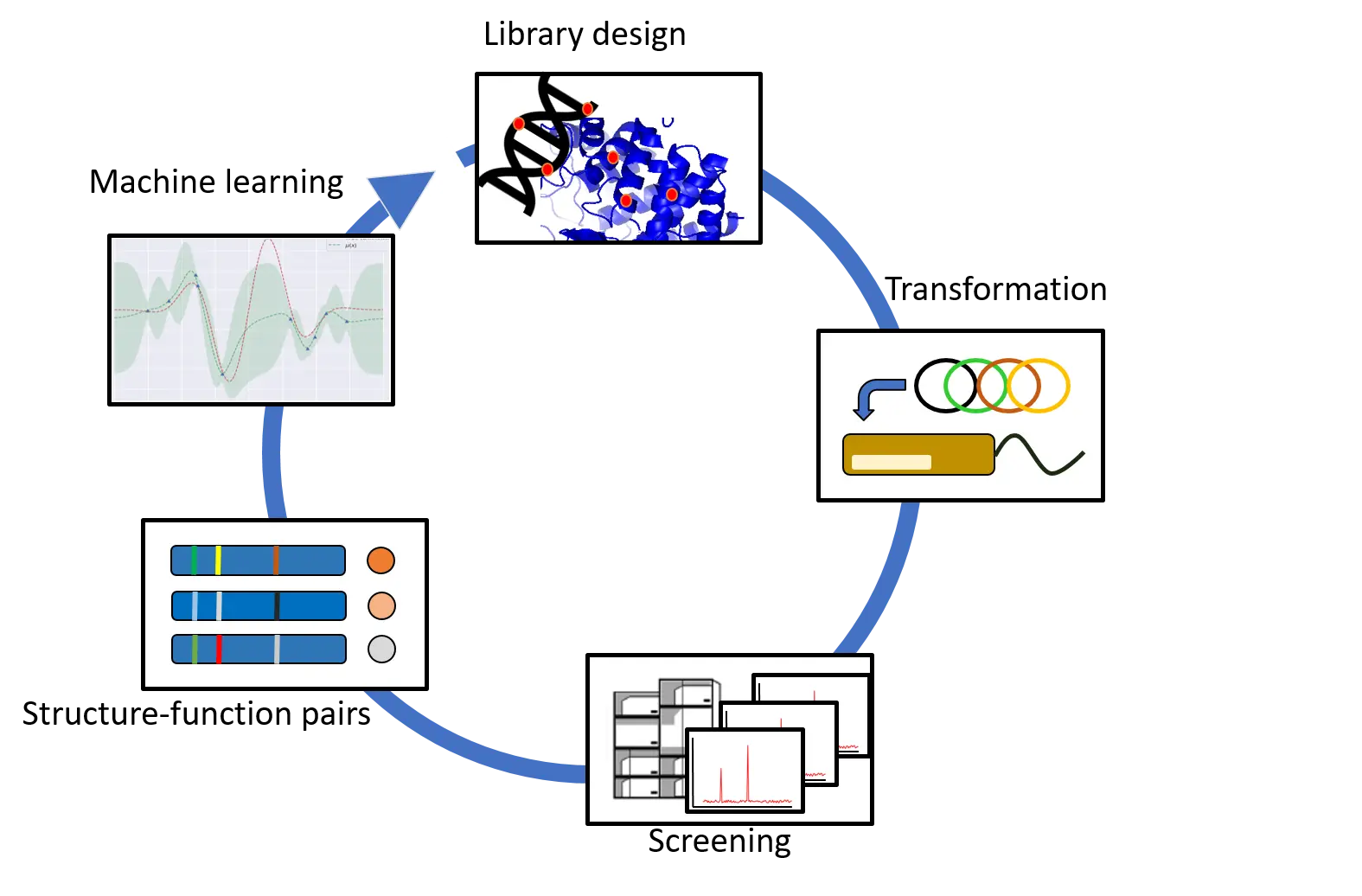
Screening large libraries is a significant bottleneck in the evolution of proteins. We are exploring the feasibility of using machine learning to identify more active variants in the vast protein space by learning sequence to function relationships on a small subset of all possible combinations.
Related Publications
- Honda Malca et al., 2024 Commun. Chem. 7: 46
- Buller et al, (2023) SCIENCE 382, 899
- Patsch et al., 2023 Comput. Struct. Biotechnol. J. 21, 4488
- Giger & Buller, 2023 CHIMIA 77, 395
- Patsch & Buller 2023 CHIMIA 77, 116
- Büchler et al., 2022 Helv. Chim. Acta 106, e202200128
- Voss et al. 2022 ChemCatChem, e202201115
- Büchler et al., 2022 Nat. Commun. 13: 371
Roboter-assisted enzyme-engineering

The screening of large enzyme libraries is very labor-intensive and thus represents a significant bottleneck in the optimization of proteins. Our customized robotic platform allows us to automate the screening workflow, from growing the relevant bacterial strains to performing the biocatalytic assays.
Related Publications
- Honda Malca et al., 2024 Commun. Chem. 7: 46
- Buller et al, (2023) SCIENCE 382, 899
- Patsch et al., 2023 Comput. Struct. Biotechnol. J. 21, 4488
- Büchler et al., 2022 Helv. Chim. Acta 106, e202200128
Enzyme Families
α-Ketoglutarate-dependent Oxygenases
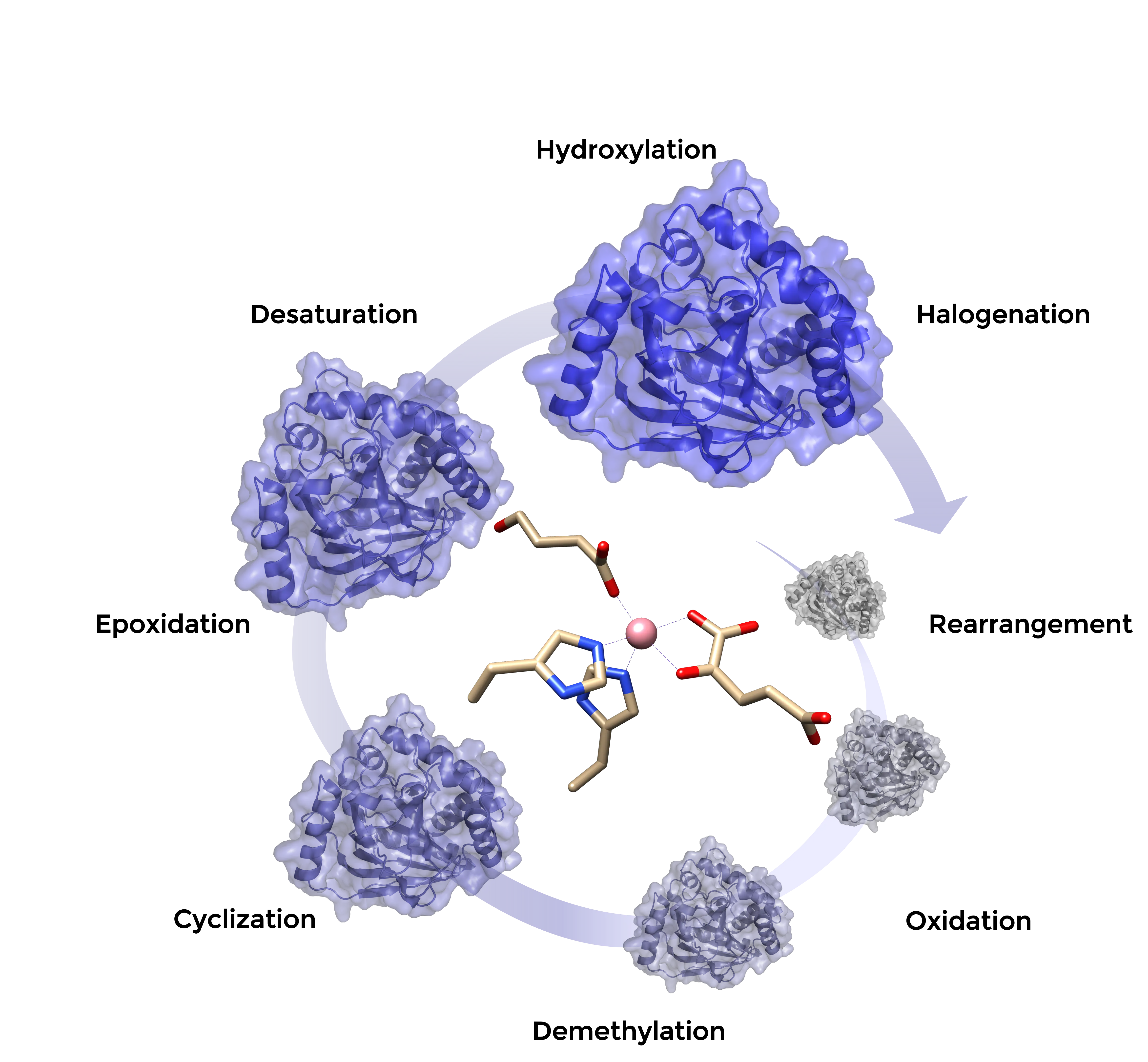
Modification of unactivated C-H bonds often challenges organic synthesis. With the aim to provide a versatile biocatalytic C-H functionalization toolbox, we explore the biocatalytic potential of Fe(II)/2-oxoglutarate dependent oxygenases. This enzyme family catalyzes a diverse set of biologically important reactions including hydroxylations, desaturations and oxidations at the expense of the co-factor 2-oxoglutarate. To broaden the enzyme’s applicability, we apply enzyme engineering to develop suitable green biocatalysts able to perform desired modifications on valuable non-native substrates.
Related publications
- Honda Malca et al., 2024 Commun. Chem. 7: 46
- Papadopoulou et a., 2022 Biochemistry 62, 229
- Büchler et al., 2022 Nat. Commun. 13: 371
- Meyer et al. 2021 ACS Catal. 11: 6261
- Voss et al. 2021 ChemCatChem 13: 1
- Frey et al., 2019 Curr. Opin. Biotechnol. 60: 29
- Peters et al., 2019 Catalysts 9: 221
Halogenases
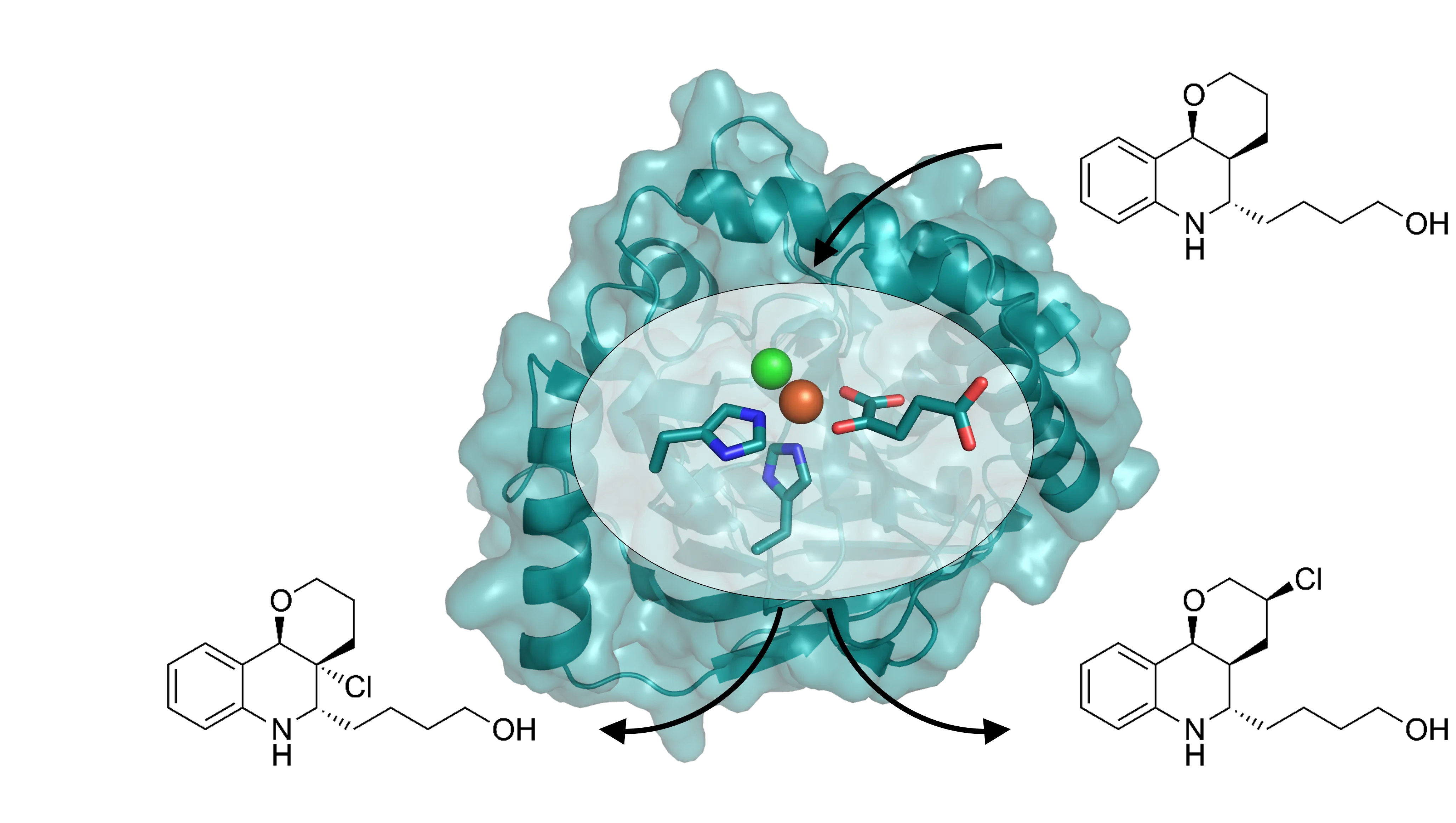
Halogenation is a transformation not often explored for industrial processes due to typically difficult-to-handle reaction conditions. Biocatalytic halogenations thus offer an elegant avenue toward the safe and selective installation of halogen atoms into bioactive molecules. Current halogenating enzymes, however, very often suffer from instability and a narrow substrate scope. We address these drawbacks by enzyme engineering and thus alter the selected enzymes’ properties in order to prepare robust and broad-use biocatalysts for aliphatic and aromatic halogenations for academic and industrial use.
Related publications
- Patsch & Buller Chimia 77, 116
- Hegarty et al., 2023 Curr. Opin. Green Sustain. Chem. 41, 100786
- Büchler et al., 2022 Helv. Chim. Acta 106, e202200128
- Voss et al. 2022 ChemCatChem, e202201115Hegarty
- Papadopoulou et a., 2022 Biochemistry 62, 229
- Büchler et al., 2022 Nat. Commun. 13: 371
- Papdopoulou et al. 2021 ChemCatChem 13: 3914
- Voss et al. 2021 ChemCatChem 13: 1
- Voss et al. 2020 Chem. Eur. J. 26: 7336
- Hayashi et al., 2019 Angew. Chem. 58: 18535
- Buechler et al. 2019 Catalysts 9: 1030
- Frey et al., 2019 Curr. Opin. Biotechnol. 60: 29
Squalene-Hopene Cyclases
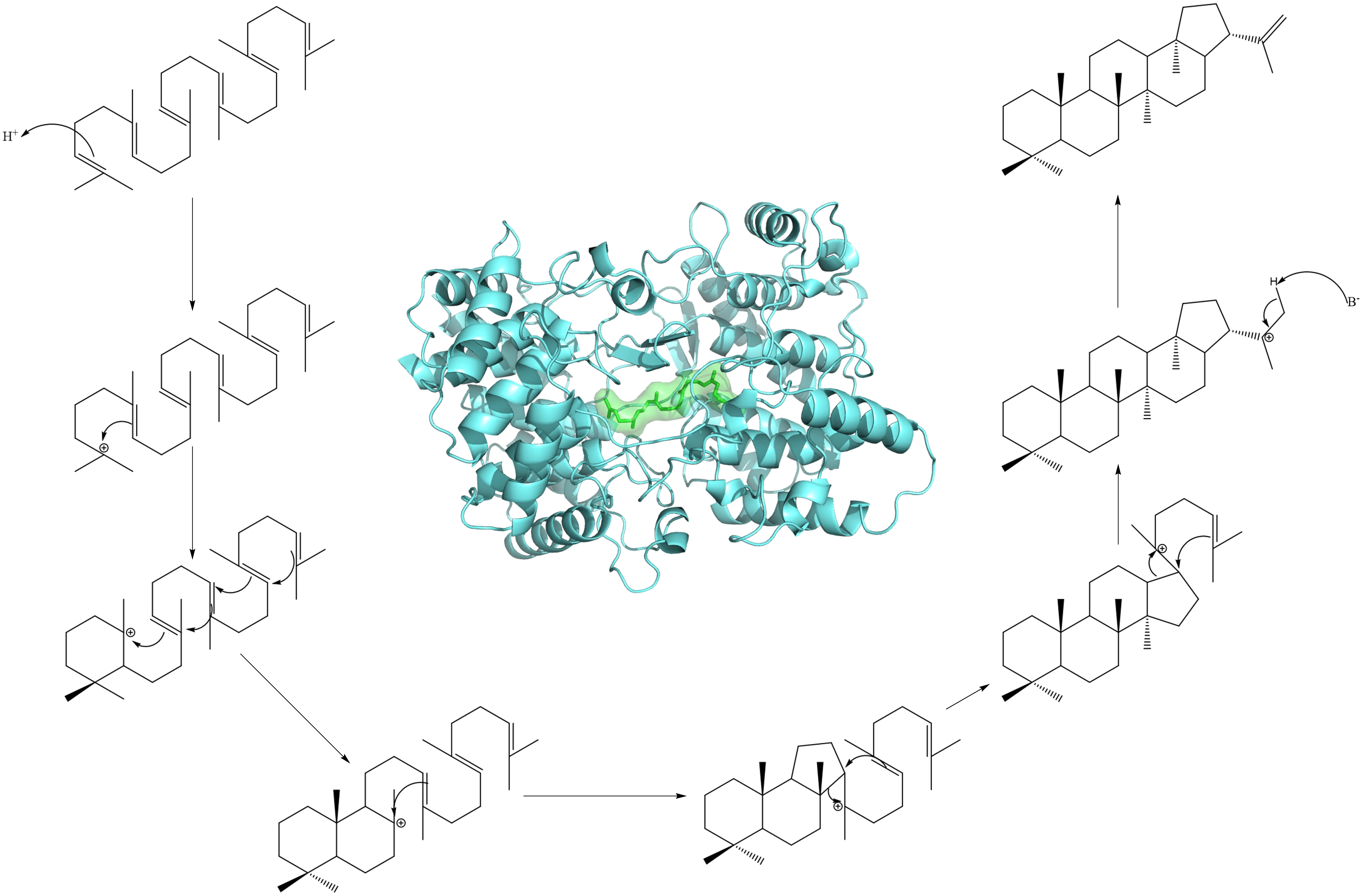
SHCs catalyze one of the most complex biochemical reactions, the cyclisation of squalene to hopene and hopanol by incorporation of five novel C-C bonds. Furthermore, these enzymes were found to exhibit a large degree of promiscuity towards industrially relevant conversions in the flavor and fragrance segment. We have generated a diverse toolbox of SHCs including novel and/or thermophilic enzymes and are currently exploring the substrate and reaction scope of these enzymes and are further optimizing them by directed evolution.
Related publications
- Patsch et al., 2023 Comput. Struct. Biotechnol. J. 21, 4488
- Eichenberger et al. 2021 Angew. Chem. 60: 26080
- Peters et al., 2019 Z. Naturforsch. C 74: 63
Ene-Reductases

Ene-reductases catalyze the asymmetric reduction of activated C=C bonds generating up to two new stereo centers. Our toolbox of ene-reductases is constantly increasing and tested for modification of chemical compounds of interest for the chemical industry.
Related publications:
- Papadopoulou et al., 2022 Org. Process. Res. Dev. 26, 2102-2110
- Aregger et al., 2020 Catalysts 10: 254
- Peters et al., 2019 ChemBioChem 20: 1569
- Peters et al., 2019 Z. Naturforsch. C 74: 63
Microbial Epimerases
Antibiotic resistant bacteria are on the rise and pose a major threat to global health. Antimicrobial peptides are promising compounds to tackle this crisis, but their clinical application is limited because of rapid degradation. (Bio)chemical modifications can result in more stable and active peptides circumventing these drawbacks. Together with Rémy Bruggmann and Vincent Perreten (University of Bern), we are developing a toolbox of enzymatic epimerases capable of carrying out specific peptide modifications.
PETase
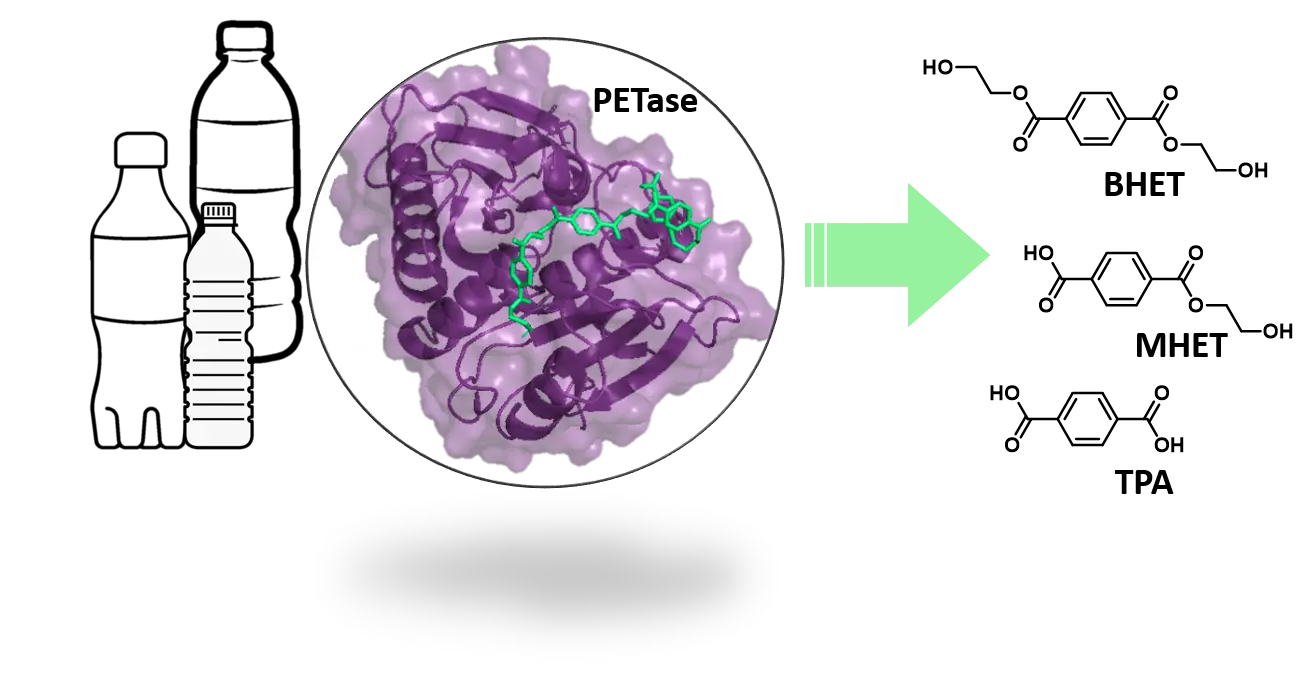
Plastic has become an indispensable part of our everyday life. Its increased production and use, however, accelerates the accumulation of plastic waste, a burden on the environment and human health. Enzymatic PET degradation has emerged as a green alternative in PET recycling and, if further improved, may become a more widely applicable strategy for general plastic waste management.
Related publication:
- Papadopoulou et al., 2019 Chimia 73: 743