Silent pathway awakening to discover novel antibacterial compounds from actinomycetes
Researchers
Prof. Dr. Martin Sievers, Prof. Dr. Rainer Riedl, David Frasson, Dr. Joël Pothier, Dr. Fabio Rezzonico, Fabienne Arn, Dr. Ivana Kroslakova, Nicola Rhyner, Irene Wüthrich (ETHZ), Steven Schmitt (ETHZ)

Description
Actinomycetes are a large group of gram-positive bacteria with a filamentous and branching growth pattern. Under certain conditions, some species produce bioactive substances such as antibiotics. Drugs produced by actinomycetes are used to treat bacterial and fungal infections as well as cancer.
One of the first antibiotics discovered from actinomycetes was streptomycin. It is produced by Streptomyces griseus, which was isolated from a farm soil at the Rutgers Agriculture School in New Brunswick, NJ and was used to treat tuberculosis. In 1952, Selman Waksman received the Nobel Prize for this work.
Most antibiotics currently being developed are modifications of existing compounds. The discovery of new classes is difficult, time-consuming, and resource-intensive. We need novel drugs quickly to combat the global antimicrobial resistance crisis and to be prepared to treat bacterial and fungal infections in the future.
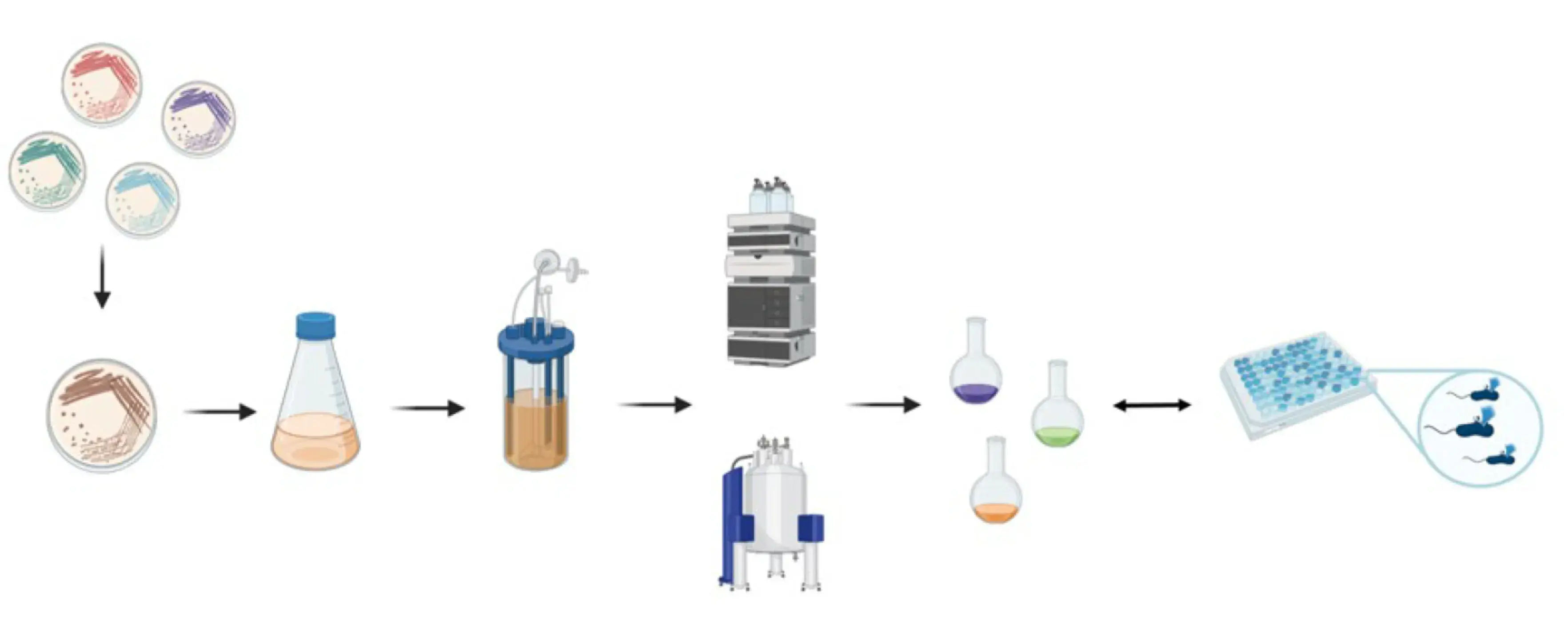
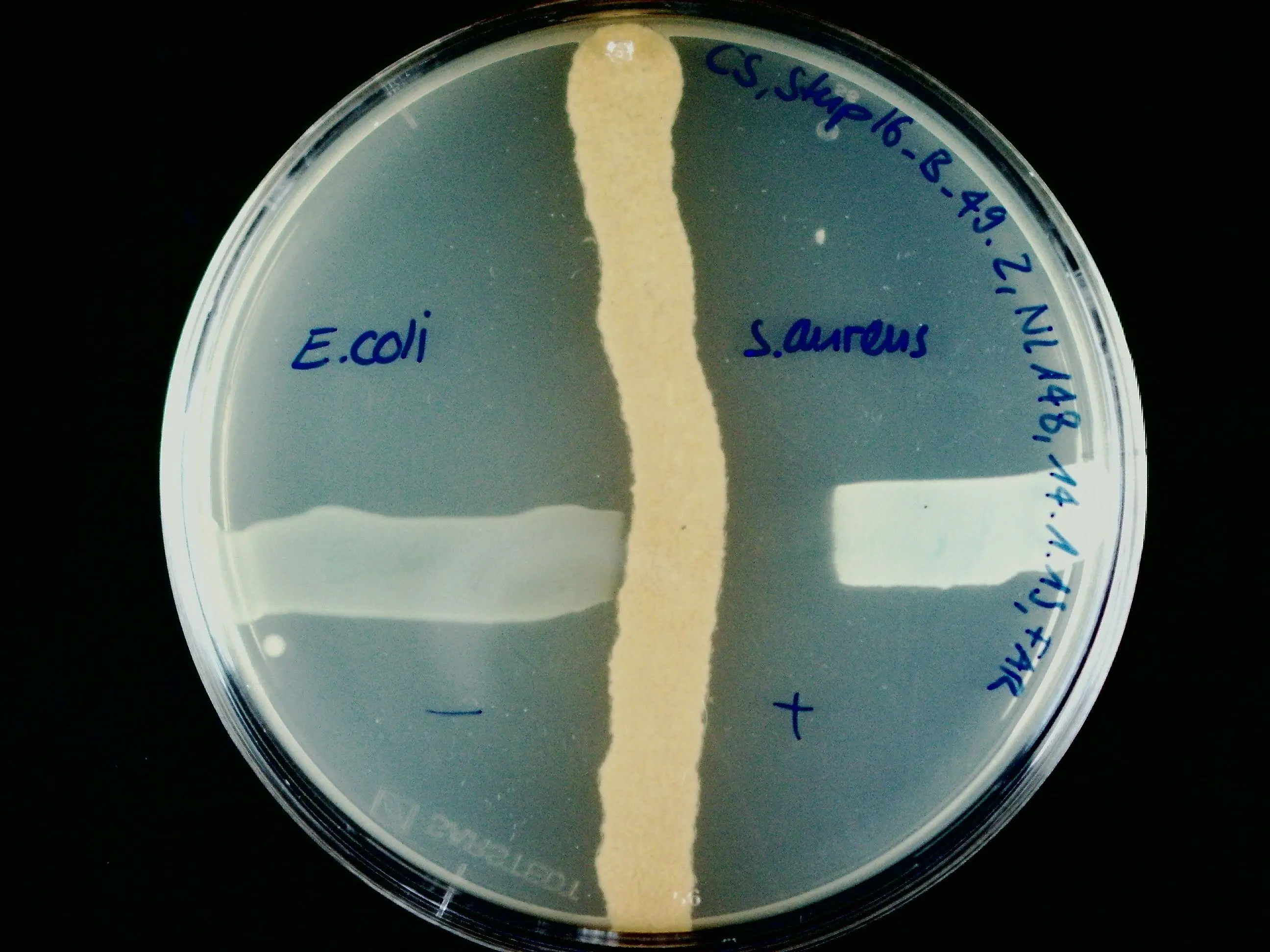
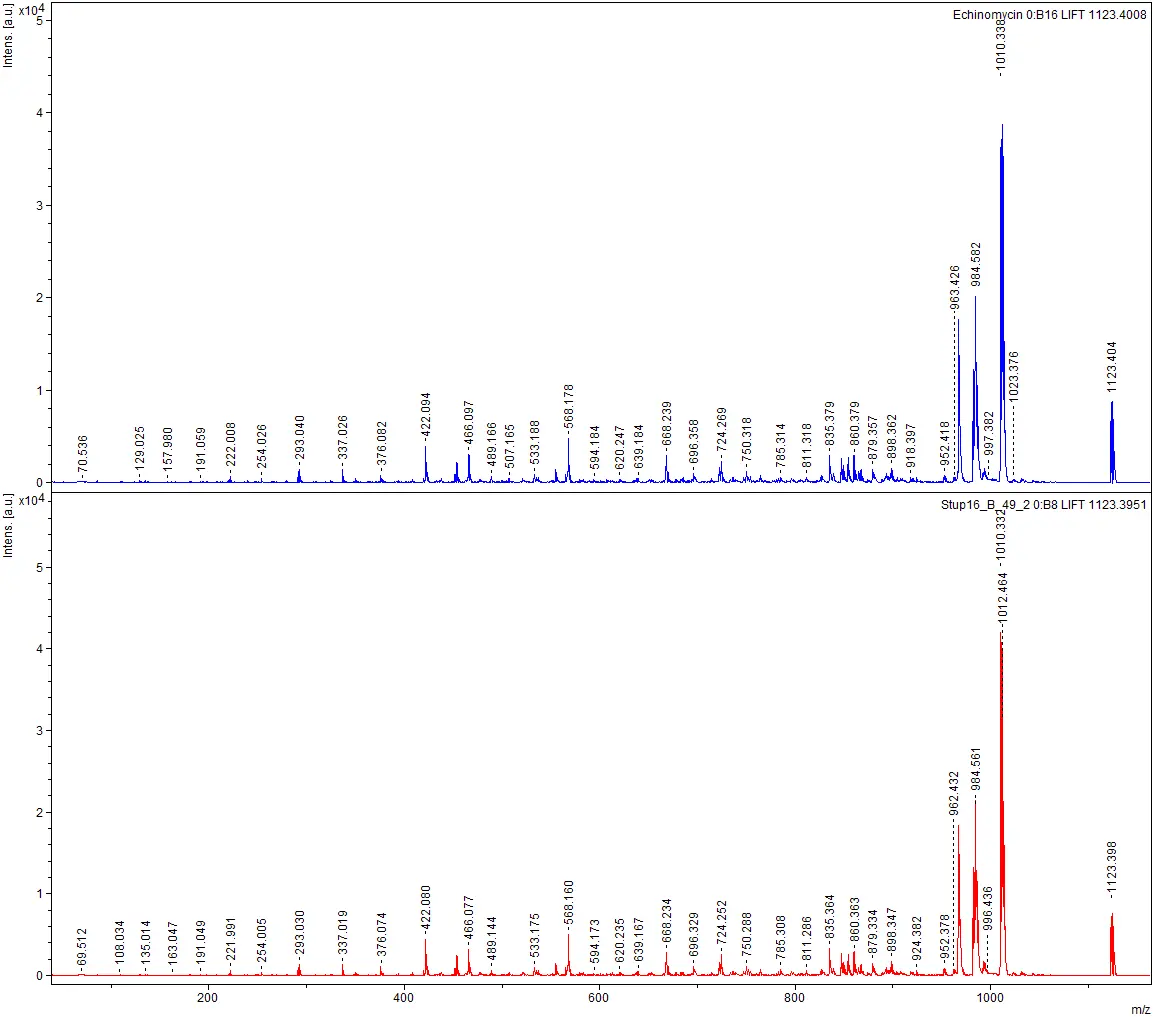
The institutes ICBT and IUNR at ZHAW developed a technology platform that combines microbial cultivation techniques, chemical analysis, and genome sequencing to identify antibiotics produced by strains of actinomycetes (Figure 1 and 2). Antibiotics like actinomycin, azalomycine, milbemycin, and echinomycin were isolated from different Streptomyces strains.
However, most actinomycetes strains isolated from their habitat silence their relevant metabolic activity and do not produce antibiotics under standard cultivation conditions in the laboratory. Therefore, novel strategies need to be developed to find new antibacterial compounds and to increase the speed of drug discovery.
In this project, we will collectively address the challenge to activate the production of novel compounds from actinomycetes. This includes the cultivation of selected actinomycetes strains directly in their natural environment using the microsphere technology platform from our external partner at ETH Zurich, as well as genomic promoter replacement to achieve constitutively expression of genes from actinomycetes encoding new antibiotics. After structure elucidation of novel antibiotics, we will furthermore establish structure-activity relationships in order to optimize their medicinal chemistry profiles toward clinical use, which is the next critical step towards the development of antibiotic drugs.