Sifting through the Data Jungle at Hospitals with AI
ZHAW researchers present their work at NeurIPS'24, one of the most impactful AI conferences worldwide. Their research focuses on electronic health records (EHRs) and querying them in natural language across various database systems, yielding new insights and solutions for the complex task of translating natural language into a query language for databases.

A major challenge in the healthcare sector and other fields is that information is stored in multiple different database architectures. For example, data can be organized in tables (relational database systems), graph databases, or document stores. To access specific information, query languages like SQL or SPARQL are used. Two graduates of the Data Science program at the ZHAW School of Engineering, Sithursan Sivasubramaniam and Cedric Osei-Akoto, developed a new multimodal solution under the supervision of Jonathan Fürst to compare different systems and query languages in terms of their AI readiness.
Their work, which was published in collaboration with ZHAW researchers Yi Zhang and Kurt Stockinger, answers for the first time the question of how well large language models (LLMs) like ChatGPT, Llama3, or Gemini can translate natural language into the respective database language. The research is presented at the renowned NeurIPS conference in Vancouver.
First Benchmark for Evaluating different Text-to-Query Systems
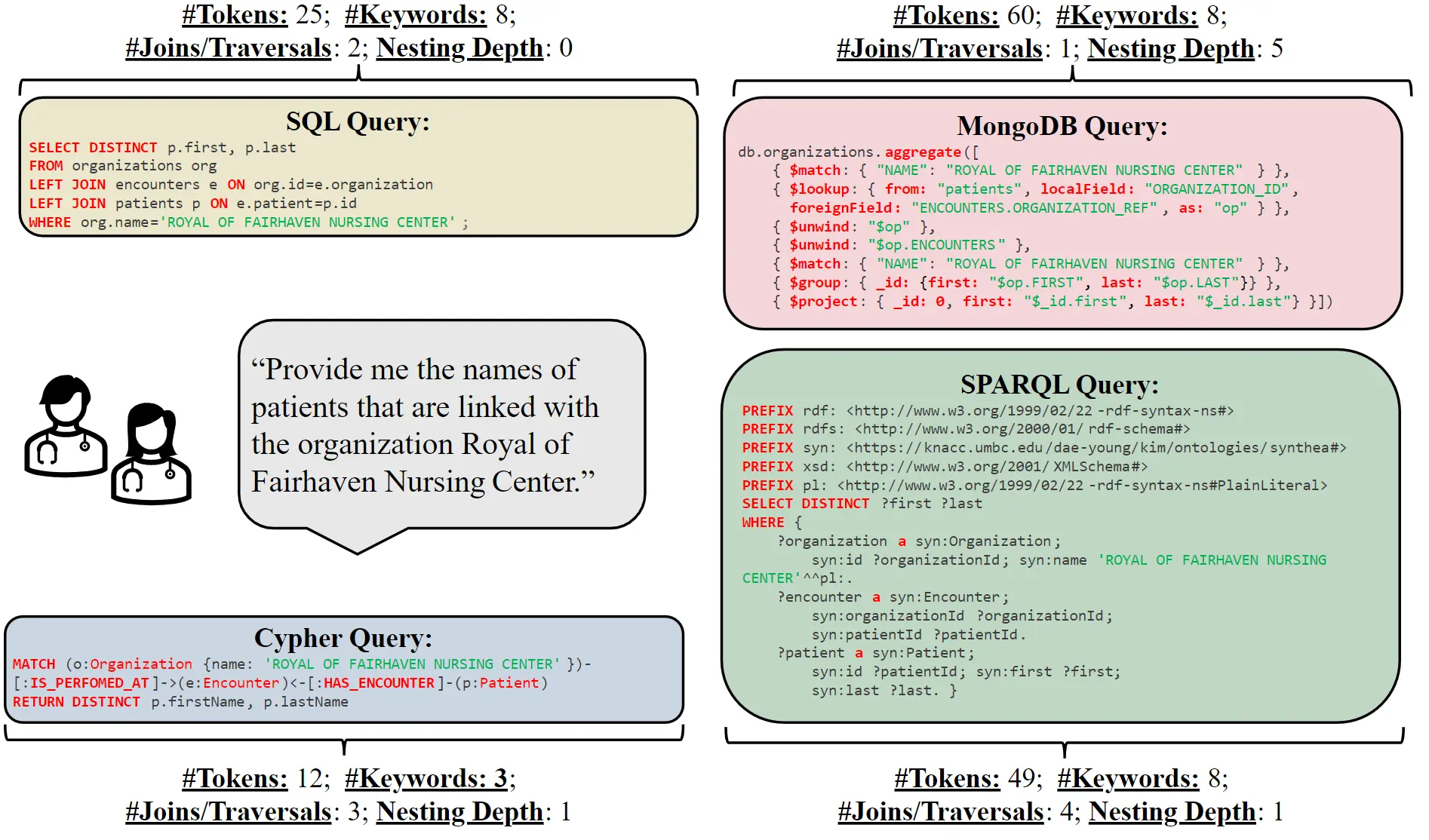
The core of the research is the SM3-Text-to-Query Benchmark, which integrates multiple database architectures and query languages for the first time. The benchmark's name is derived from the three "Ms" (multi-modal and medical). Text-to-Query means that a text in natural language triggers a query in the database, e.g., "Find patient names with symptoms ABC in hospital XYZ." Based on synthetic, i.e. automatically generated and therefore data protection-compliant, patient data and a comprehensive health terminology (SNOMED-CT taxonomy), this benchmark supports various database architectures, enabling a comprehensive evaluation of the performance of different systems.
Innovative Methodology and Comprehensive Evaluation
The researchers developed 408 question templates, which were expanded into a comprehensive benchmark set of 10,000 natural language/query pairs for each query language. These query languages include four well-known languages (SQL, MQL, Cypher, and SPARQL). The goal of the experiments with these data was to determine how well large language models (LLMs) can translate natural language into these four different query languages. Two open (both LLama from Meta) and two closed language models (Gemini from Google and ChatGPT from OpenAI) were analyzed.
Results and Outlook
The study's results show significant differences in the performance of the various database models and query languages. In the future, the benchmark is expected to be adapted to additional query languages or real, standard-based patient databases, making it a valuable tool for future research and development in the field of digital healthcare. With their work, the researchers set new standards in the evaluation of text-to-query systems. This standardised and privacy-preserving benchmark provides the scientific community and the general public with valuable insights and solutions to the challenges of modern healthcare.
More information
- NeurIPS Conference: 2024 Conference
- Paper: https://doi.org/10.48550/arXiv.2411.05521
- Code and data: https://github.com/jf87/SM3-Text-to-Query