Forschungsgruppe Biomedical Simulation

Introduction
It is truly ambitious to model natural processes. Nature is unique and highly complex, modelling in turn is per definition reductionist.
Cells converse with other cells via sending proteins signals. On a micro-scale many cells form functional compounds and integrate into complex structures often embedded in a matrix. The proteins in turn are regulated by genes, which may be regulated by macroscopic factors like mechanical stimuli. The cell is a typical example where the micro and macro world are closely intertwined. It is our mission to better understand these complex interactions.
Systems biology combines biology with simulations and develops effective validation strategies. Simulating here in essence means: gaining insights while managing the unknown.
The motivations for modelling has many facets, which are often tiered:
- to understand a system and its boundaries via replicating its behaviour,
- to optimize scenarios or to characterize a system by parameter fitting,
- to predict the system behaviour under altered environmental conditions.
Sven Hirsch über das Forschungsprojekt AneuX (Gehirnaneurysmas)

Projects & services
Modelling of complex systems
System dynamics is a methodology to simulate and understand the evolution of systems. A system may be as profane as a swinging pendulum or as complex as a biochemical process. The basic concept is simple, yet universal: controlling interconnected buckets and flows. Translated into maths this means coupled ordinary and partial differential equations. This mechanistic theory unfolds into a highly versatile toolset to mimic real world situations and understand their behaviour. We have applied this technique to chemical process optimization, logistics simulations, bioreactor optimization, traffic system planning and market forecast to name but a few.
Our system dynamics approach allows us to build efficiently robust models. Apt visualization tools facilitate the interpretation of the simulation results.
Multi physics simulation
Bridges, motors and airplanes are nowadays simulated with so-called continuum formulations like finite element methods (FEM). Geometrical objects are subdivided into sufficiently small elements (e.g. triangles or tetrahedra) to capture spatial effects. Between these elements interactions are formulated using partial differential equations.
This structural mechanic approach can be applied to other physical theories like fluid flow, heat transfer, chemical reactions, diffusion or sound wave propagation. Multi physics means to simulate various physical effects at the same time, e.g. fluid flow with heat transfer, or an exothermal chemical mixing reaction.
We have simulated heat propagation in various foods to determine food safety constraints. In the medical domain we have developed a new simulation approach to predict blood flow and blood coagulation, tumour growth or the remodelling of cerebral aneurysm. Applications can also be as pragmatic as computational fluid dynamics simulations of stent deployment.
Modelling of biological and medical systems
Biological systems reveal peculiarities such as evolution and natural variability. Still, the inherent variability of organic systems can be accounted for leading to predictable behaviours. This is the domain of systems biology: it is a constant communication effort between scientific disciplines. In systems biology we combine our broad modelling expertise and analytical vigour with in depth process understanding.
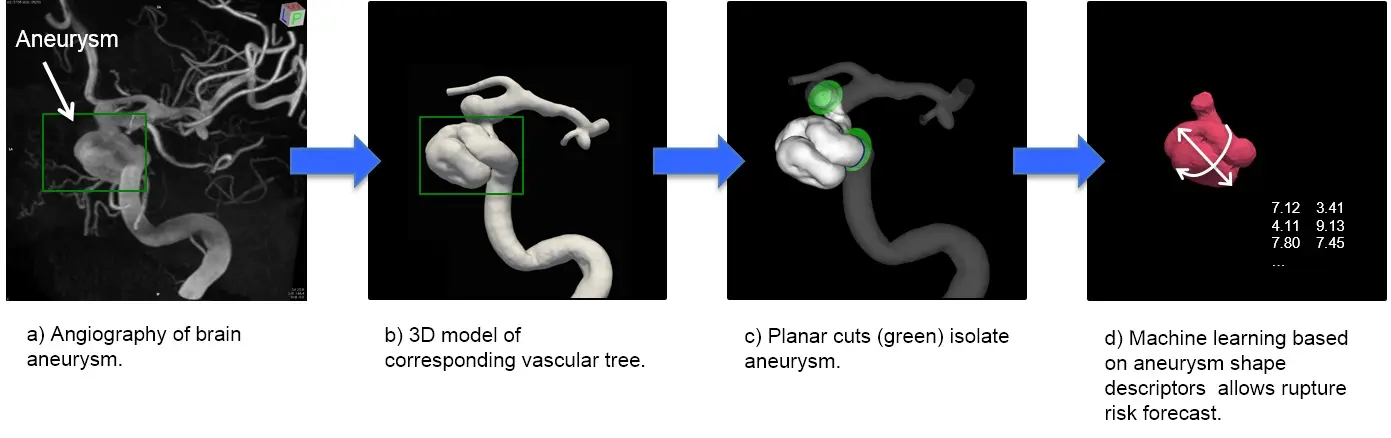
Cerebral aneurysm and vascular biology are currently our focal interests. A cerebral aneurysm is a pathological bulging of the brain artery. Its rupture is potentially deadly. We are investigating the wall shear stress sensing within blood vessels and their effect on aneurysm shape and growth. Our team of basic scientists and computer scientists collaborates with clinicians and biologists to understand the role of an aneurysm’s shape on its risk to rupture. Here we use machine learning approaches to interpret the data (Figure 1).
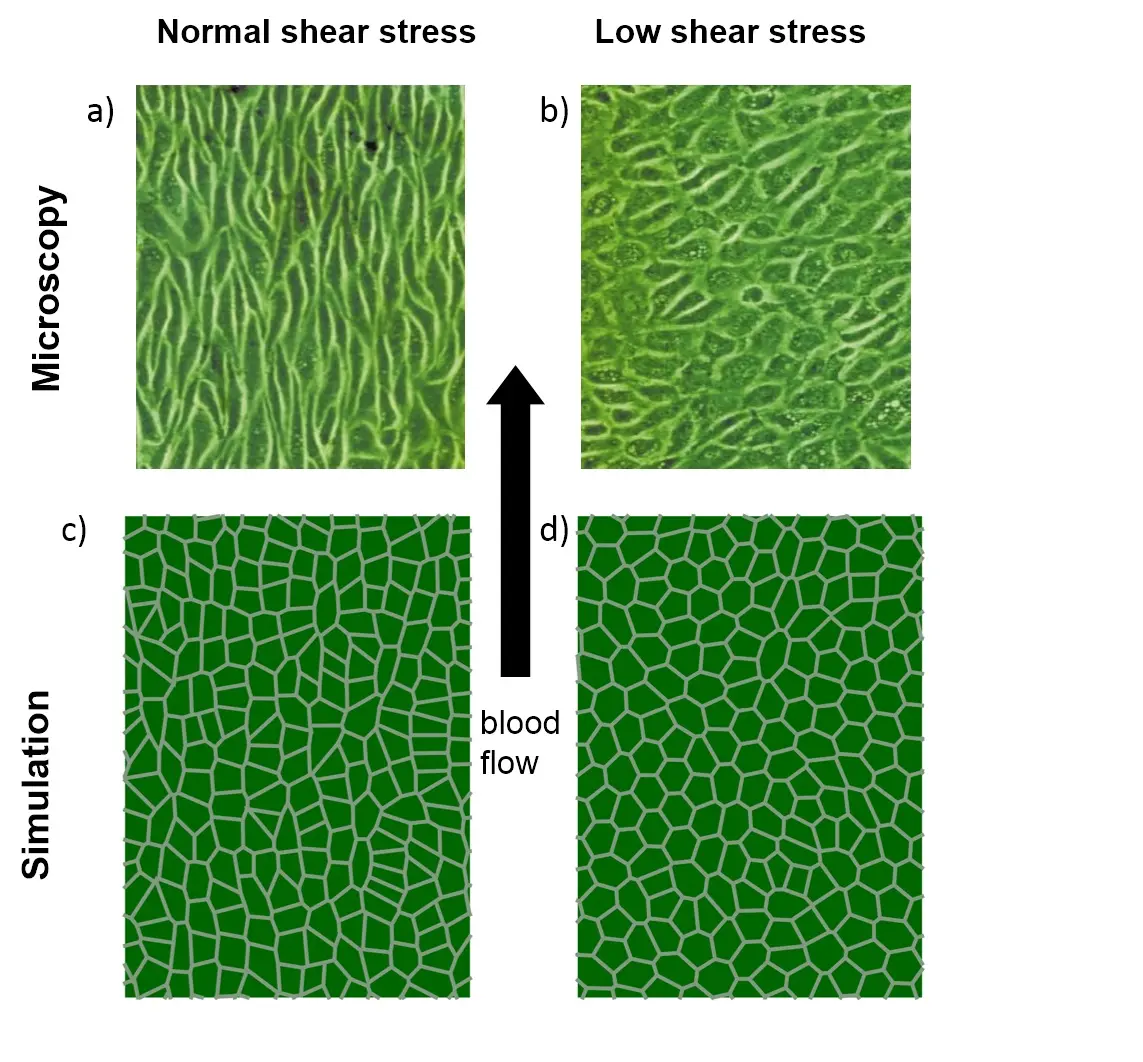
Modelling biological systems means to be able to flexibly adapt our research strategies to accommodate the very question. Often we are extending or combining established modelling approaches. If advised we will develop our own approaches. Figure 2 shows such an example: We developed a cell vertex models to understand the change in the shape of endothelial cells under different arterial blood flow conditions.
Many of our applications have a direct clinical impact. Typically we are addressing basic research questions where translation is already anticipated. We are dedicated to bridge the gap from basic science to delivering tangible results.
Reference project: AneuX, supported by SystemsX
Links / Partners
University of Zurich, Interface group (Prof. Vartan Kurtuoglu)
CABMM – Center of applied Biotechnology and Molecular Medicine
Read more about the Institute's network
Team
-

ZHAW Life Sciences und Facility Management
Schloss
8820 Wädenswil -

ZHAW Life Sciences und Facility Management
Schloss
8820 Wädenswil